Accuracy and data efficiency in deep learning models of protein expression
15 Dec 2022· ,,,·
1 min read
,,,·
1 min read
Evangelos-Marios Nikolados
Arin Wongprommoon
Oisin Mac Aodha
Guillaume Cambray
Diego A. Oyarzún
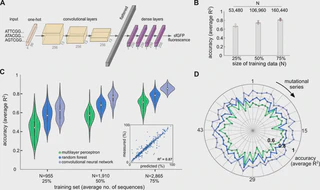 Image credit: [Nikolados et al.]
Image credit: [Nikolados et al.]Abstract
Synthetic biology often involves engineering microbial strains to express high-value proteins. Thanks to progress in rapid DNA synthesis and sequencing, deep learning has emerged as a promising approach to build sequence-to-expression models for strain optimization. But such models need large and costly training data that create steep entry barriers for many laboratories. Here we study the relation between accuracy and data efficiency in an atlas of machine learning models trained on datasets of varied size and sequence diversity. We show that deep learning can achieve good prediction accuracy with much smaller datasets than previously thought. We demonstrate that controlled sequence diversity leads to substantial gains in data efficiency and employed Explainable AI to show that convolutional neural networks can finely discriminate between input DNA sequences. Our results provide guidelines for designing genotype-phenotype screens that balance cost and quality of training data, thus helping promote the wider adoption of deep learning in the biotechnology sector.
Type
Publication
Nature Communications
Lay summary
Deep learning, a type of machine learning, can be used to predict how much protein a specific DNA sequence can produce. But, it usually needs a lot of data.
We managed to get a deep learning model to make predictions with less data, especially if the data is diverse.
Our study is useful because people can then produce fewer DNA to train a model, and this is cheaper.
My role
I made sure that the Python code for the non-deep and deep learning models was robust and easy for people to use.
I also gave feedback on the manuscript. My comments improved a figure; it shows that a diverse DNA sequence input improved how efficiently the model worked and how much it was able to handle a variety of sequences.